An article titled “m6A RNA modification as a new player in R-loop regulation,” was published in the January 2020 edition of Nature Genetics and widely reported in the scientific community. What we learn from it opens the door for crucially important knowledge in the context of this technology being used for Covid vaccines.
We just found out that Facebook made this information illegal on its platform and cancelled a whole field of science, while Fauci denied its existence, all knowing the truth is different. Praise Veritas!
UPDATE #2: I’ve just unearthed a 2017 Ted Talk featuring the current Moderna boss Tal Zaks, where he describes the mRNA technology that was first meant to treat cancer, he calls it “information therapy”, see for yourselves:
UPDATE #3: MAY 11, 2021 – Father of the Human Genome Project crushes Fauci’s and Zuckerberg’s stupid lies with his new publicly available technology
Below I copy/pasted the press release that circulated at the time in several top publications:
“Following a new collaboration between UiO and research groups in Nottingham and Oxford, it has now been revealed that RNA has a direct effect on DNA stability, according to Professor Klungland’s research.
He believes the discovery will provide the health service with an important tool, since many studies have shown that the regulation of modifications to RNA is important for the development of cancer.
If genes that are important for the chemical compound 6-methyladenine are completely removed, this results in neurodegeneration in both mice and humans.
Where and how
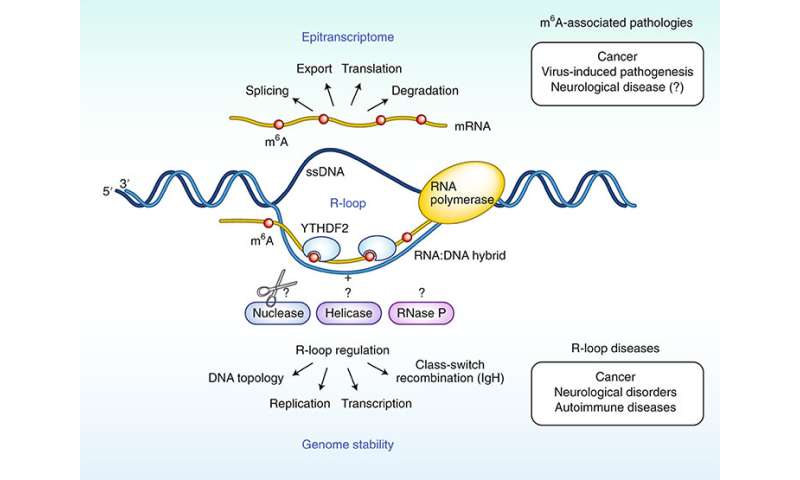
In areas of DNA where RNA binds to one of the DNA threads in such a way that the complementary DNA thread becomes the sole thread (R-loop structures), the DNA stability will change if RNA is chemically modified by m6A.
“Several research groups are now working together to study what effect this can have on the DNA molecule. We already know that R-loop areas are associated with sequences of DNA containing active genes and that this can lead to chromosomal breakage and the loss of genetic information”
Prof. Arne Klungland
New field of research
Normally, epigenetic gene regulation is studied by examining dynamic modifications of DNA and proteins—so-called epigenetic modifications. The modifications can turn genes on or off without changing the underlying genetic code.
Less than 10 years ago, it was discovered that dynamic modifications also exist in RNA and that these have an important role to play in gene regulation
Important modification
The most common modification is on mRNA is 6-metyladenin (m6A). It has now been shown that this modification is essential for the survival of cells and model (non-human) organisms.
Over the last five years, there has been an enormous increase in the amount of research into RNA modifications—a field called epitranscriptomics.
One of the first studies in this field of research was the result of a collaboration between research groups in Chicago, Beijing and Oslo (Zheng, Dahl et al., Molecular Cell, 2012, 49, 18-29).
End of article citation.
I bolded a few paragraphs to make sure you see what I saw:
RNA modification not only can alter DNA, but it’s been already envisioned as a tool for DNA editing.
This argues against the whole BS official narrative that the RNA vaccine technology is inoffensive for the DNA
One of the most remarkable findings of this study is that depletion of YTHDF2 and METTL3 (the writer that deposits m6A) increases levels of γH2AX, a marker of DNA double-strand breaks, thus suggesting that pathological R-loop accumulation in the absence of the m6A RNA-methylation pathway challenges genome integrity. This result is in line with findings from many studies that clearly suggest the potential of R-loops to induce DNA double-strand breaks2. Moreover, dysregulation of R-loops is emerging as a critical factor driving genome instability in a large variety of pathological contexts, including after oncogenic stress12, in cells infected with Kaposi’s sarcoma–associated herpesvirus, in neurological disorders associated with trinucleotide-repeat expansion (such as Huntington’s disease and fragile X syndrome) and in multiple other inherited ataxias (for example, ataxia with oculomotor apraxia 2)2. The use of existing drugs targeting the m6A pathway (for example, inhibitors of FTO) could therefore be considered as a new therapeutic approach to treat R-loop-related diseases13.
m6A RNA modification as a new player in R-loop regulation – Aline Marnef &
Beyond genome stability, the finding that m6A methylation controls R-loop levels across the genome considerably expands the biological functions of the m6A-modification pathway to potentially all R-loop-related functions, including DNA topology (because R-loops have recently been proposed to relieve superhelical stress), immunoglobulin class-switch recombination (and therefore the immune response), replication initiation and transcription1,2,14. Additionally, m6A accumulates at sites of ultraviolet-induced DNA damage6, thus raising the interesting possibility that this modification may also regulate R-loops during DNA repair and thus affect the frequency of chromosomal translocations15.
In summary, the results presented by Abakir et al. unveil an unexpected interplay between RNA modifications (the epitranscriptome) and the maintenance of genome integrity. Re-analysis of the pathological contexts implicating dysregulation of the m6A RNA pathway through the prism of genome instability therefore warrants further investigation (Fig. 1).
Gaëlle Legube, Nature Genetics

Epitranscriptomics: The new RNA code and the race to drug it
A small group of scientists studying chemical modifications on RNA ushered in the field of epitranscriptomics. Now they’re hoping it will create an entirely new way to treat cancer
by Ryan Cross, Chemical & Engineering News, Feb. 18, 2019
It’s not every day that a biotech investor stumbles across an entirely new field of science. And frankly, Carlo Rizzuto wasn’t even looking for such a thing. When Rizzuto, a partner at the venture capital firm Versant Ventures, embarked on a scouting trip to New York City in 2014, he was simply hoping to discover academic research that was ripe enough to form the basis of a biotech company.
Rizzuto had an appointment with Samie Jaffrey, an RNA scientist at Weill Cornell Medicine. RNA is often described as a cousin to DNA—the stuff that our genes are made of. One kind of RNA, called messenger RNA, acts as the intermediary code that cells use to transfer information stored in DNA into a set of instructions that cells can easily read for making proteins.
After Rizzuto rejected several of his projects, Jaffrey mentioned a relatively young line of work focused on studying chemical modifications to RNA. In 2012, his lab invented a method to map the location of methyl groups that, for some reason, cells were adding to their mRNA. It was reminiscent of another field, called epigenetics, or the study of chemical modifications made to DNA to turn genes on or off. The entirety of RNA in a cell is called the transcriptome, so Jaffrey dubbed the new field “epitranscriptomics.”
Rizzuto perked up. “This is something that we would be very interested in,” he said. Credit: Gotham TherapeuticsSamie Jaffrey, a professor of pharmacology at Weill Cornell Medicine and cofounder of Gotham Therapeutics, explains the m6A modification on RNA. Jaffrey’s lab invented a technique to map the location of m6A on RNA.
Credit: Gotham TherapeuticsSamie Jaffrey, a professor of pharmacology at Weill Cornell Medicine and cofounder of Gotham Therapeutics, explains the m6A modification on RNA. Jaffrey’s lab invented a technique to map the location of m6A on RNA.
Jaffrey was hesitant. “We’re just doing basic stuff now,” he recalls explaining. His lab, and others, was still trying to figure out how this RNA modification system worked. They were building evidence suggesting that enzymes added and removed these methyl marks to control the fate of mRNA, and thus protein production, but many questions remained. Jaffrey implored: “Carlo, what disease would we be curing if we started a company around epitranscriptomics?”
“It doesn’t matter,” Rizzuto replied. “This is so central to molecular biology; it has to be related to fundamental disease processes.”
Then reality kicked in. Venture capital firms like Rizzuto’s aren’t in the business of funding years of basic research just to see if something like epitranscriptomics is involved in disease. “We were looking at a new paradigm for gene-expression regulation,” Rizzuto recalls, but it was too early to start a company. He and Jaffrey agreed to stay in touch.
Rizzuto’s enthusiasm in 2014 has since percolated among scientists and investors learning about epitranscriptomics. Several groups, including Jaffrey’s, have shown that the epitranscriptomic code—the number and location of chemical modifications across a cell’s RNA—is seriously out of whack in some cancers. And with basic tools in hand to read this previously hidden layer of information in cells, biotech companies are now out to alter it. Three start-ups, including one that Jaffrey and Rizzuto helped found, called Gotham Therapeutics, have launched with more than $110 million in total dedicated to epitranscriptomics drug discovery.
There was a similar reaction to epigenetics more than a decade ago, when it became clear that chemical modifications regulating genes are frequently out of whack in cancer. Companies rushed to develop drugs against proteins responsible for making, removing, and recognizing chemical modifications on genes—often referred to as the writer, eraser, and reader proteins. With the discovery of parallel writer, eraser, and reader proteins working on RNA, epitranscriptomics is looking like a promising, untapped area for drug discovery.
But there’s another parallel to epigenetics that’s less optimistic: thus far, epigenetic drugs have been a disappointment. “Epigenetics turned out to be a lot more complicated than the community originally thought,” says Chuan He, a professor of chemistry at the University of Chicago.
He, a scientific founder of the epitranscriptomics company Accent Therapeutics, has been at the forefront of developing the new study of RNA modifications and their role in disease. He, Jaffrey, and many others are confident that understanding and controlling RNA modifications will provide completely new avenues for treating disease. “What this really offers is a totally new biology,” He says. “And whenever there is a new biology emerging there are always opportunities for therapies.”
EPITRANSCRIPTOMICS, ABRIDGED
A series of discoveries and technical advancements over the past decade has spawned a new field called epitranscriptomics, the study of chemical modifications to RNA, and the proteins that write, erase, and read these modifications. In recent years, studies implicating epitranscriptomic proteins in cancer have led to the launch of three biotech companies dedicated to drugging these proteins.
May 2008: Rupert Fray shows that a methyl-adding enzyme is essential for plant development. The study inspires others to look at RNA modifications.
November 2010: Chuan He proposes new field of RNA epigenetics, suggesting that methyl modifications on RNA can be removed.
October 2011: Chuan He’s lab proves that an enzyme called FTO erases methyl modifications on RNA.
April and May 2012: The labs of Gideon Rechavi (April) and Samie Jaffrey (May) publish the first maps of RNA methyl modifications. Jaffrey coins the word “epitranscriptomics.”
October 2014: Howard Chang’s lab shows that METTL3, which adds methyl groups to RNA, is critical for embryonic stem cell development and differentiation.
June 2016: Storm Therapeutics, founded by University of Cambridge scientists Tony Kouzarides and Eric Miska, raises $16 million to drug proteins that make RNA modifications.
September and November 2017: Independent studies from Samie Jaffrey and colleagues (September) and Tony Kouzarides and colleagues (November) show that METTL3 is elevated in acute myeloid leukemia and that suppressing the enzyme forces the cancer cells to become noncancerous.
May 2018: Accent Therapeutics, cofounded by Chuan He, Howard Chang, and Robert Copeland, raises $40 million.
October 2018: Gotham Therapeutics, cofounded by Samie Jaffrey, launches with $54 million.
February 2019: Evidence builds that epitranscriptomics may be important for cancer immunotherapy. Chuan He shows that deleting a reader protein boosts the efficacy of checkpoint inhibitors in mice.
MAKING A MAP
A series of events beginning in 2008 laid the foundation for epitranscriptomics. That year, while He was studying epigenetic enzymes that remove methyl modifications from DNA, he and University of Chicago biologist Tao Pan began doubting that all these enzymes were really working on DNA as others assumed. The evidence was particularly shaky for one enzyme, called fat mass and obesity-associated protein, or FTO.
But a study coming out of the lab of plant biologist Rupert Fray at the University of Nottingham reinforced He and Pan’s suspicions that RNA modifications were underappreciated. Fray showed that plants missing a methyl-adding enzyme—similar to an enzyme called METTL3 in humans—stopped growing at a specific early stage in their development.
Scientists knew that METTL3 placed a methyl on a specific nitrogen in adenosine, one of the four building blocks of RNA. This modified building block is called N6-methyladenosine, or m6A for short. Beyond m6A, chemists had cataloged some 150 different chemical modifications to RNA in bacteria, plants, and animals. If He could find an enzyme that removed the methyl groups, it would suggest that there was an undiscovered RNA control system in cells, analogous to epigenetic controls in DNA.
In 2010, He coined the phrase “RNA epigenetics” in a commentary that outlined his ideas (Nat. Chem. Biol., DOI: 10.1038/nchembio.482). A year later, He and Pan published evidence showing that the FTO enzyme was an eraser—it removed the methyl modifications made by METTL3 (Nat. Chem. Biol. 2011, DOI: 10.1038/nchembio.687).
METTL3 and FTO are both enzymes, which means they should be pretty straightforward to inhibit with small-molecule drugs. That notion would later be frequently cited by the new epitranscriptomics companies, although it would be several years still before these enzymes were connected to disease.
At first, the significance of these enzymes was lost on many researchers. At Weill Cornell, however, Jaffrey immediately recognized that He’s study was part of a new field that was about to explode. His lab had been working on a method to detect and map m6A across a cell’s mRNA. Jaffrey had also seen Fray’s work on m6A in plants and thought that if the modifications existed in humans, they must be doing something important in us too.
At the time, methods for studying m6A were rudimentary. Researchers could detect the presence of m6A in ground-up globs of mRNA run through common chemistry lab techniques like chromatography or mass spectrometry. “But you had no idea which mRNAs were being modified,” Jaffrey says. No one knew if all mRNA had some m6A or if the methyl modifications were found on only certain transcripts, he adds. “And frankly, it wasn’t even terribly clear that m6A levels changed.”This is so central to molecular biology; it has to be related to fundamental disease processes.Carlo Rizzuto, partner, Versant Ventures
So Jaffrey and Kate Meyer, a postdoc in his lab, developed a technique to figure out which mRNAs contained these modifications. They used commercially available antibodies that attach to m6A to fish out fragments of human mRNA for sequencing (Cell 2012, DOI: 10.1016/j.cell.2012.05.003).
That technique allowed the creation of the first map of m6A. The results were stunning. “We thought that m6A was going to be all over the place, kind of random,” Jaffrey says. Instead, the researchers saw that methyl marks tended to cluster near an area called the stop codon, and only on certain mRNA transcripts. “It was so specific, it just knocked our socks off.”
An even closer inspection revealed that many of the mRNAs containing m6A were linked to differentiation and development, the same functions that were affected in Fray’s stunted plant embryos. “We were amazed,” Jaffrey says.
In April 2012, while Jaffrey and Meyer were waiting for their m6A paper to publish, another group, led by Gideon Rechavi at Tel Aviv University, published its own paper on the use of antibodies to map m6A in mouse and human cells (Nature 2012, DOI: 10.1038/nature11112). “It was met with a lot of skepticism,” says Dan Dominissini, the PhD student in Rechavi’s lab who led the project. “People didn’t get why it was important. It took a year to publish.”
The problem was researchers still hadn’t established a clear link between these RNA modifications and disease, or even basic human biology. Moreover, the field wouldn’t have its name of epitranscriptomics for another three weeks, when Jaffrey and Meyer’s paper describing their m6A-mapping technique was published online in May 2012. Although Jaffrey had been scooped, the back-to-back publications put epitranscriptomics on the radar. The field was poised to explode.
Editing the epitranscriptomic code
The most common RNA modification is N6-methyladenosine (m6A), which is made when a protein complex containing the “writer” enzyme METTL3 adds a methyl group to adenosine. Two different “eraser” enzymes, called ALKBH5 and FTO, can remove a methyl group to turn m6A back into adenosine.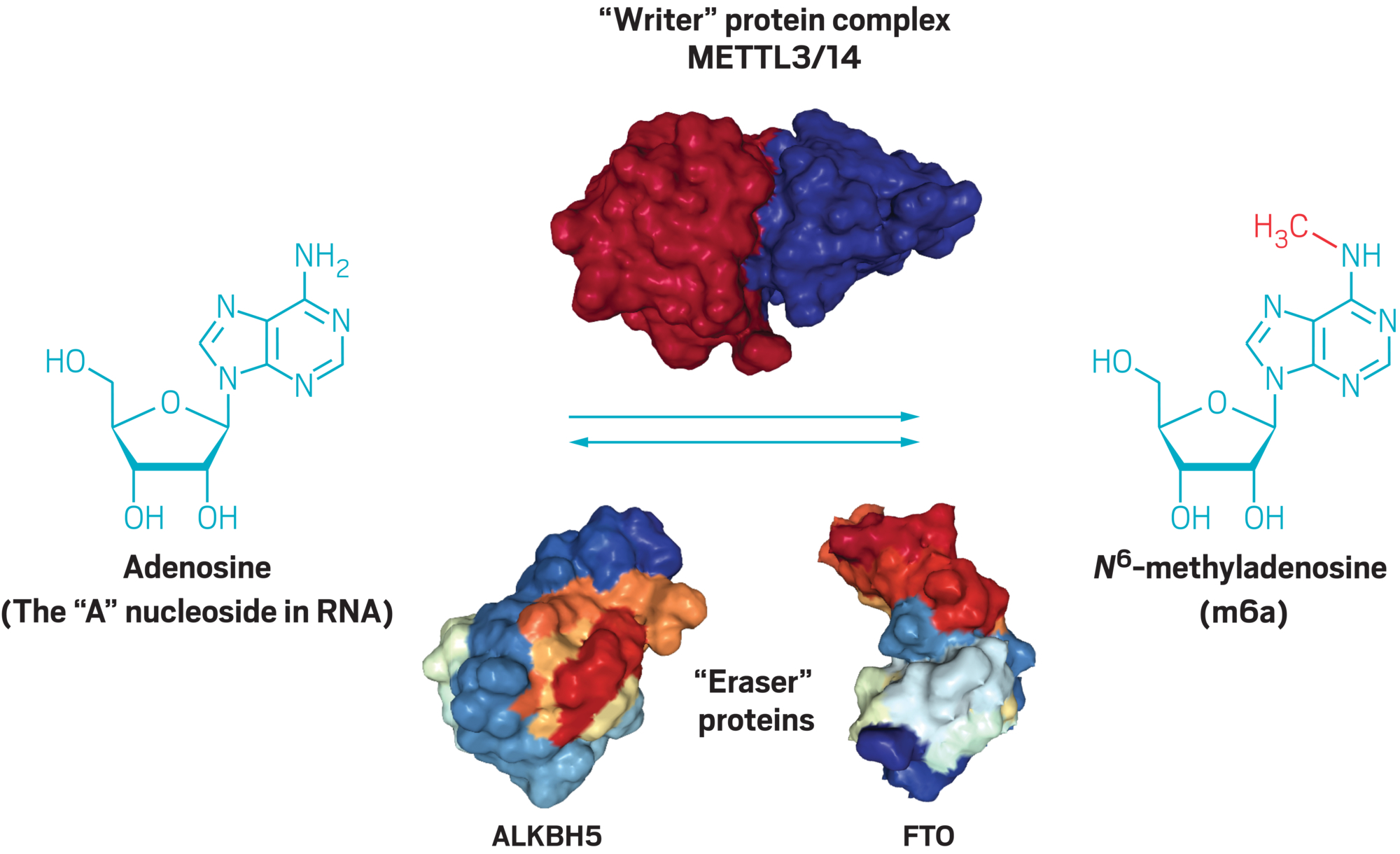 Credit: ALKBH5, FTO, and METTL3-METTL14 protein images created with the Protein Data Bank, NGL Viewer
Credit: ALKBH5, FTO, and METTL3-METTL14 protein images created with the Protein Data Bank, NGL Viewer
SEARCHING FOR DISEASE
In Chicago, He was positioning his lab as the forefront of epitranscriptomics research. His group discovered that an enzyme called ALKBH5, like FTO, erased methyl marks on RNA, turning m6A back into adenosine. Yet even by 2014, two years after the m6A-mapping methods were published, epitranscriptomics wasn’t getting the recognition, or funding, that He thought it deserved. “People thought it was cute,” He says. “But biologists were not convinced of its significance.”
Epitranscriptomics was now a hot topic. As studies began bubbling up exploring the role of RNA modifications, particularly m6A, in a variety of cells and species, investors started putting money into the field. In June 2016, a British start-up called Storm Therapeutics raised $16 million and became the first company dedicated to tackling the new RNA epigenetics.
Although Storm was several years in the making, it wasn’t clear what diseases the company would be curing. Two University of Cambridge scientists, Tony Kouzarides and Eric Miska, began discussing the idea for the company back in 2012, when they had published work on obscure enzymes that chemically modify microRNAs, which regulate the function of other RNAs.
Although the enzymes were linked to cancer, at least in cells growing in a dish, the microRNA studies went largely unnoticed. Kouzarides and Miska thought more undiscovered links between RNA modifications and cancer must exist, but it took a few years to find investors willing to bet on their hypothesis. “I don’t think that there was a huge amount of actual data; it was just the belief that there must be,” Storm’s CEO, Keith Blundy, says. “The idea that all of these chemical modifications on RNA weren’t dysregulated or mutated or changed in cancer was almost unthinkable.”
That belief, which echoes the sentiment that Versant Ventures’ Rizzuto expressed in Jaffrey’s office in 2014, was about to be validated. In the second half of 2016, studies began linking reader and writer proteins to cancer. Jaffrey saw the evidence firsthand in an ongoing study he was conducting in blood cancer. The implications for drug discovery were becoming clear. He reached out to Rizzuto. It was time to move forward.
ANNOTATIONS IN THE BLOOD
The common thread running through epitranscriptomics research was its link to cell differentiation and development. Chang’s and Rechavi’s stem cell studies on m6A gave several research labs—including He’s, Jaffrey’s, and Kouzarides’s—the idea to look at the role of these RNA modifications in a deadly blood cancer called acute myeloid leukemia.
Leukemia is essentially a disease of dysfunctional differentiation. Healthy people’s bones are filled with hematopoietic stem cells that produce white blood cells. In leukemia, these stem cells go haywire. They proliferate and displace other blood cells because they can’t differentiate, or mature, into normal white blood cells.
In December 2016, He’s lab, together with several collaborators, showed that tissue samples taken from people with certain kinds of acute myeloid leukemia displayed high levels of the enzyme FTO—which, five years earlier, He had discovered is an m6A eraser (Cancer Cell 2016, DOI: 10.1016/j.ccell.2016.11.017). A few months later, with a different set of collaborators, He showed that levels of the methyl-removing enzyme ALKBH5 were elevated in glioblastoma stem cells (Cancer Cell 2017, DOI: 10.1016/j.ccell.2017.02.013).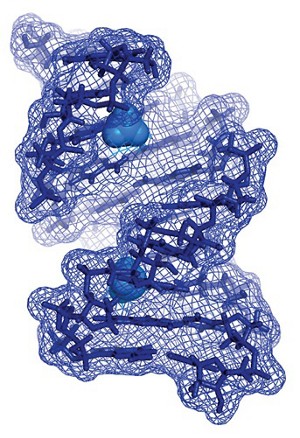 Credit: Journal of the American Chemical SocietyA surface (mesh) structure of an RNA duplex (sticks) with the methyl modification of m6A (balls).
Credit: Journal of the American Chemical SocietyA surface (mesh) structure of an RNA duplex (sticks) with the methyl modification of m6A (balls).
At the beginning of 2017, Lasky, the Column Group investor, reached out to He. Now that epitranscriptomic enzymes were tied to cancer, Lasky’s firm wanted to start a drug company to control RNA modifications. With the new cancer data in hand, He felt that the time was right.
The investors also knew about a publication in the works from Jaffrey and leukemia expert Michael Kharas at Memorial Sloan Kettering Cancer Center. The Column Group and Versant Ventures worked together for a time to begin forming a single epitranscriptomics company with several of the academic leaders. During the summer of 2017 however, the different players split into two camps. The Column Group brought on He and Chang as academic cofounders of Accent Therapeutics. Versant Ventures named Jaffrey the academic founder of Gotham Therapeutics.
While Accent and Gotham were still in stealth mode, Jaffrey published a study showing that genetic mutations led to fixed, elevated levels of METTL3 in acute myeloid leukemia, keeping white blood cells from forming. By reducing METTL3 levels, leukemia cells could be coaxed into undergoing differentiation to become noncancerous cells that eventually die (Nat. Med. 2017, DOI: 10.1038/nm.4416). “It was remarkable because we didn’t even need complete inhibition of METTL3,” Jaffrey says.
Two months later, Kouzarides’s lab at the University of Cambridge published similar results, with additional details on what METTL3 was doing in these cells (Nature 2017, DOI: 10.1038/nature24678). In leukemia, elevated METTL3 encouraged the production of proteins linked to cancer. “It is feeding the cell the very proteins that are driving tumorigenesis,” Gotham CEO Lee Babiss says.
Epitranscriptomics now had drug targets, diseases, and high-profile studies. After recruiting additional investors, Accent launched with $40 million in May 2018, and Gotham launched with $54 million in October. Storm Therapeutics is in the process of raising approximately $65 million for its second round of cash from investors. Although none of these companies will name their targets or first diseases they will attempt to treat, conversations with the companies’ CEOs suggest that developing inhibitors of METTL3 is a goal for all three.
Drug designers have a lot of experience inhibiting enzymes, making METTL3 an attractive first target. But its activity may not be straightforward, says Yunsun Nam, a biophysicist at the University of Texas Southwestern Medical Center. METTL3 grabs the methyl group it adds to RNA from S-adenosylmethionine (SAM), a molecule used by several other enzymes. Companies’ compounds will need to avoid inhibiting these other enzymes as well, she explains.
Nam thinks a workaround could be targeting a protein called METTL14, which is attached to METTL3 as part of a larger m6A-writing complex. “METTL3 and METTL14 are very dependent on each other for stability,” she says.
Even if the companies can develop selective METTL3 inhibitors, it’s unclear how many people would benefit from them. While the leukemia studies by Jaffrey and Kouzarides showed that m6A levels are too high, He’s leukemia and glioblastoma studies showed the opposite, that m6A levels are too low. Other studies have suggested more contradictory results—including that m6A levels may be too high in glioblastoma. In other words, when developing therapies, it will be crucial to know the epitranscriptomic state of one’s cancer cells. Otherwise, giving the wrong person a METTL3 inhibitor might make things worse.
“That’s a possibility,” Robert Copeland, the president and chief scientific officer of Accent, acknowledges. The challenge for Accent and other companies will be to figure out which subset of people with leukemia would benefit from a METTL3 inhibitor, to lower m6A levels, and which would benefit from an FTO inhibitor, to raise m6A levels, Copeland explains. “If the pendulum swings too much one way or too much the other way, you can cause disease.” Credit: Accent TherapeuticsRobert Copeland, president and chief scientific officer of Accent Therapeutics
Credit: Accent TherapeuticsRobert Copeland, president and chief scientific officer of Accent Therapeutics
EPI-EXPANSION
Although the leaders of Accent, Gotham, and Storm are being secretive about their strategies, they all hint that the potential scope of epitranscriptomics drug discovery is much bigger than just targeting METTL3.
In addition to the m6A erasers, a growing body of work is uncovering the importance of the m6A readers. Earlier this month, He’s lab showed that an m6A reader protein called YTHDF1 is an important control switch in the immune system and that inhibiting it might dramatically boost the efficacy of existing checkpoint inhibitors, a popular class of cancer immunotherapy (Nature 2019, DOI: 10.1038/s41586-019-0916-x). “I think a lot of immunotherapy companies will jump into epitranscriptomics once they read the paper,” He says.
And this isn’t the first known link between epitranscriptomics and immunotherapy, Accent’s Copeland says. His firm has been studying an enzyme called ADAR1—which stands for adenosine deaminase acting on RNA—that modifies adenosine bases in RNA. Studies from academic labs show that some tumors depend on ADAR1 in ways that normal cells do not. One study suggests that blocking ADAR1 could make certain drug-resistant cancers vulnerable to checkpoint inhibitors (Nature 2018, DOI: 10.1038/s41586-018-0768-9).
Other labs entering the fray are uncovering new proteins that read, write, and erase RNA modifications, with links to additional types of cancer and other diseases. The scope of epitranscriptomics could be enormous. “That’s what excites us about the field,” says Blundy, Storm’s CEO. “There are many, many RNA pathways that are regulated through modifications.”
The discoveries aren’t all coming smoothly, however. For example, Jaffrey claims that the main target of the eraser enzyme FTO isn’t actually m6A but a slightly different modification, called m6Am. Others disagree. “There is still some debate, but that is the normal trajectory for a field, especially in the early days,” Jaffrey says.
The field also still has technical hurdles. “Right now the methods to map and detect m6A are crude,” Jaffrey admits. Existing methods require large sample sizes and are ineffective at quantifying how m6A levels change over time on particular mRNA transcripts. His lab is now working on ways to better quantify m6A to diagnose or predict diseases in the clinic. Such tools will be critical for recruiting the right people into clinical studies testing inhibitors of epitranscriptomic proteins.
Another issue is the lack of publicly available small-molecule inhibitors for studying epitranscriptomic proteins. “We don’t even have an inhibitor for research,” says Dominissini, who has also developed new RNA-modification-mapping techniques and now runs his own epitranscriptomics lab at Tel Aviv University. Right now, researchers have to use genetic techniques to remove or block production of writer, eraser, and reader proteins, but what the field really needs are simple small molecules to test the hypotheses that these proteins will make good drug targets, he says. Of course, that’s what the companies are working on.
A similar lack of compounds stalled epigenetics drug discovery more than a decade ago. Pioneers in the epitranscriptomics field are unfazed by these parallels. “I don’t think there is a relationship between the success or failure of an epigenetics drug to an epitranscriptomics drug,” Jaffrey says.
The scientists and companies in the field are running full speed ahead. Hundreds of labs have cited papers from Dominissini, He, and Jaffrey, and all can point to several ongoing studies investigating the role of RNA modifications in other diseases. “It reflects how fast people jumped into the field,” He says. “Epitranscriptomics is booming.”
RNA AS A PATHWAY TO THE BRAIN
A 2018 study from the Scripps Research laboratory of Sathyanarayanan Puthanveettil, PhD, peers deep within the nucleus of developing brain cells and finds that long noncoding RNAs play an important role in the healthy functioning and maintenance of synapses, the communication points between nerve cells in the brain.
“Long noncoding RNAs are often described as ‘the dark matter of the genome.’ So, systematic interrogation of their function will illuminate molecular mechanisms of brain development, storage of long-term memories and degradation of memory during aging and dementia,” Puthanveettil says.
RNA are the master regulators of the cell, tiny chains of nucleotides that read, transcribe and regulate expression of DNA, and build proteins. While scientists have gained great insights recently into the genetics underpinning how brain cells reach out and communicate with each other, the role of noncoding RNA is poorly understood. Research suggests that the longest of these noncoding RNA, those over 200 nucleotides long, help determine which genes are activated and operating in brain cells at various times. But which ones?
Writing in the journal Proceedings of the National Academy of Sciences, Puthanveettil and his colleagues on Scripps Research’s Florida campus report that a specific long non-coding RNA, GM12371, controls expression of multiple genes involved in nervous system development and functioning. Furthermore, it affects the developing neurons’ shape and ability to signal.
In mouse hippocampal cells, learning-related signaling upregulates GM12371, while its reduction produces inactive neurons, ones with sparse branches.
Together, the results suggest that healthy growth and development of brain cells and brain circuits depends not just upon specific proteins but also upon specific long noncoding RNAs, which scientists are now beginning to explore.
What role GM12371 dysfunction may play in diseases of the brain and nervous system demands further study, Puthanveettil says.
“Both coding and noncoding RNAs are increasingly viewed as druggable targets. Identifying their specific roles in the fundamental biology of functioning of neural circuits might eventually open new ways of treating neuropsychiatric disorders, such as autism and Alzheimer’s disease,” Puthanveettil says.
A Chinese team of researchers furthers these findings one year later:
“In the emerging field of epitranscriptomic mechanisms, mRNA m6A modification has potential role in learning and memory. It regulates physiological and stress-induced behavior in the adult mammalian brain, and augments the strength of weak memories. As a newly identified element in the region-specific gene regulatory network in the mouse brain, mRNA m6A modification plays a vital role in the death of dopaminergic neuron.
“The role of mRNA m6A methylation in the nervous system”, Cell & Bioscience, 2019
Mettl3-mediated RNA m6A modification has the direct effect on regulating hippocampal-dependent long-term memory formation. The decrease of Mettl3 in the mice hippocampus may reduce its memory consolidation, and adequate training or restoration would restore the ability of learn and memory.”
From the same Chinese study quoted above:
“Epitranscriptomics, also known as “RNA epigenetics”, is a chemical modification for RNA regulation [1]. According to its function, RNA can be divided into two broad categories, including encoding protein mRNA and non-coding RNA. With the deep research of epitranscriptomics, the researchers found methylation modification on mRNA, which is involved in the regulation of eukaryotic gene expression [2,3,4].
The mRNA is a type of RNA with genetic information synthesized by DNA transcription, which acts as a template in protein synthesis and determines the amino acid sequence of the peptide chain [5]. It is an important RNA in the human body. The methylation is the process of catalytically transferring a methyl group from an active methyl compound such as S-adenosylmethionine (SAM) to another compound, which can chemically modify certain proteins or nucleic acids to form a methylated product [6]. In biological systems, methylation influences heavy metal modification, regulation of gene expression, regulation of protein function, RNA processing, etc. [7]. At the early 1970s, scientists discovered the presence of the methylation modification in mRNA [8, 9]. The mRNA methylation modification mainly located in the nitrogen atom of the base group to form m6A, which is enriched in long exons and overrepresented in transcripts with alternative splicing variants [10]. The mRNA methylation modifications also include 5-methylcytosine (m5C), N1-methyladenosine (m1A), 5-hydroxymethylcytosine (5hmC), N6, 2′-O-dimethyladenosine (m6Am), 7-methylguanine (m7G) (Fig. 1). These modifications can affect regulation of various biological processes, such as RNA stability and mRNA translation, and abnormal mRNA methylation is linked to many diseases“.
Another US study from 2018 deals with “Role of RNA modifications in brain and behavior” and reveals that:
“Much progress in our understanding of RNA metabolism has been made since the first RNA nucleoside modification was identified in 1957. Many of these modifications are found in noncoding RNAs but recent interest has focused on coding RNAs. Here, we summarize current knowledge of cellular consequences of RNA modifications, with a special emphasis on neuropsychiatric disorders. We present evidence for the existence of an “RNA code,” similar to the histone code, that fine-tunes gene expression in the nervous system by using combinations of different RNA modifications. Unlike the relatively stable genetic code, this combinatorial RNA epigenetic code, or epitranscriptome, may be dynamically reprogrammed as a cause or consequence of psychiatric disorders. We discuss potential mechanisms linking disregulation of the epitranscriptome with brain disorders and identify potential new avenues of research”.
But the most important take out from this latter study, for me, is the final conclusion that stresses the need for larger data-bases to advance the research. I find it important because data bases need samples. And samples are often collected with swabs, like those used for Covid testing.
“With the development of more and better epitranscriptome sequencing technologies there will be a need to analyze large sequencing datasets. New bioinformatic tools are needed to supplement the current data analysis pipelines which were initially designed to analyze chromatin immunoprecipitation sequencing (ChIP seq) data. These new tools will need to take into account the complications caused by differential splicing, and amplification bias induced during reverse transcription as well as integrate multiple RNA modifications within the same molecule of RNA, across the entire transcriptome. A comprehensive database for curating and sharing epitranscriptomic data should be established to standardize the experimental and computational procedures that are used in different studies.123 We envision that in the not so distant future many new molecular and bioinformatic tools will become available to facilitate rapid advancements in the field of epitranscriptomics.”
“Role of RNA modifications in brain and behavior” – Y. Jung and D. Goldman
Actually China and US have been collaborating for quite a while in getting ahead of the curve in RNA therapies. In 2017, a mixed research team from the two countries noted in a study:
“Over 100 types of chemical modifications have been identified in cellular RNAs. While the 5′ cap modification and the poly(A) tail of eukaryotic mRNA play key roles in regulation, internal modifications are gaining attention for their roles in mRNA metabolism. The most abundant internal mRNA modification is N⁶-methyladenosine (m⁶A), and identification of proteins that install, recognize, and remove this and other marks have revealed roles for mRNA modification in nearly every aspect of the mRNA life cycle, as well as in various cellular, developmental, and disease processes. Abundant noncoding RNAs such as tRNAs, rRNAs, and spliceosomal RNAs are also heavily modified and depend on the modifications for their biogenesis and function. Our understanding of the biological contributions of these different chemical modifications is beginning to take shape, but it’s clear that in both coding and noncoding RNAs, dynamic modifications represent a new layer of control of genetic information.”
Dynamic RNA Modifications in Gene Expression Regulation, 2017, Cell magazine

It’s obvious we’re dealing with an already vast scientific domain that can expand far and wide and has serious positive and negative potential for the human species.
And that’s a completely different story than what the establishment is giving you on the RNA vaccines and technologies.
In closure, I’ll quote none other than the Oxford University, the world-famous covid vaccine developers who have been also at the core of this technology, as proven by their 2018 study “m6A modification of non-coding RNA and the control of mammalian gene expression“
Conclusions
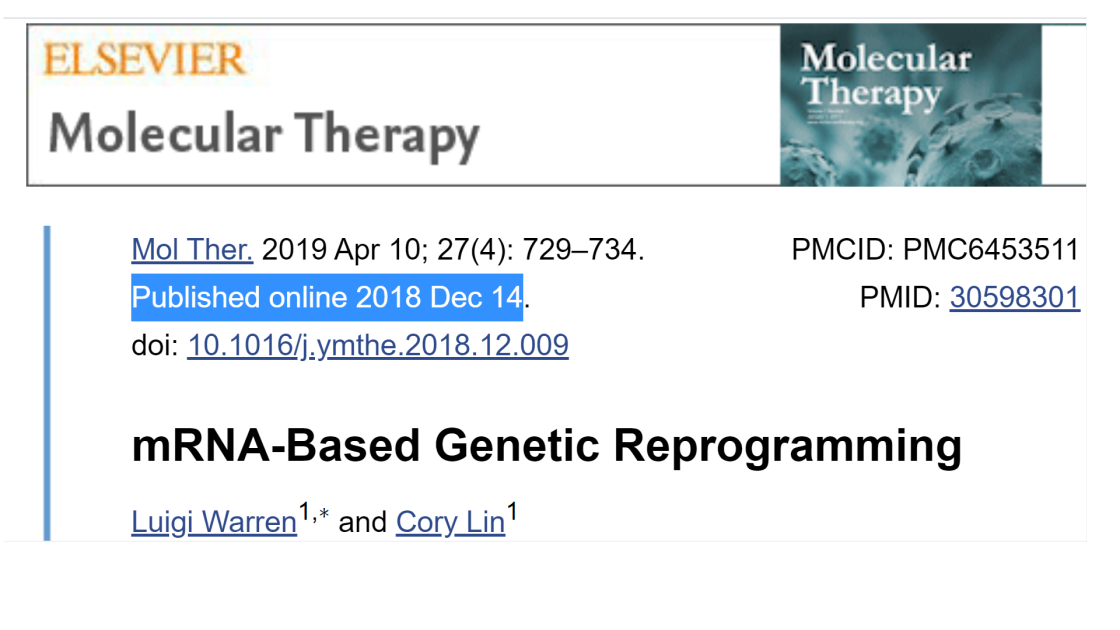
“Of the techniques so far demonstrated that can make iPSCs with useful efficiency, mRNA transfection affords the cleanest solution to the problems associated with gene expression vector persistence, obviating any need to screen for residual traces of vector and minimizing any concerns that the reprogramming system will leave an imprint on the iPSCs. From this standpoint alone, it appears to be a strong contender for application to iPSC production in the clinical arena. It also offers advantages with respect to speed and efficiency that may translate to benefits at the level of genomic integrity in a mass-production setting, assuming the polyclonal iPSC expansion strategy described above gains favor. The labor-intensive character of the original protocol, perhaps the biggest drawback to the technique, has been greatly alleviated in more recent versions of the system. The difficulty of reprogramming blood lineages with mRNA remains a significant challenge, but it is by no means clear that blood will be the starting material of choice for future clinical-grade iPSC production. In conclusion, the mRNA reprogramming system offers an attractive path around one of the main stumbling blocks to future iPSC-based therapeutics and, accordingly, continues to deserve and receive the attention of scientists working to bring that dream to reality.” – MOLECULAR THERAPY
LUIGI WARREN is founder and CEO of Cellular Reprogramming, Inc., of Pasadena, CA, an mRNA reprogramming service provider.
UPDATE:
Last minute paper from RNA Biology confirms a scientific reality that can be simplified as: RNA can be used not only as a backdoor to your DNA, but also to your brain, with the potential to make you and your future generations dumb without anyone ever suspecting it. We don’t know if this is being currently done, but we have the tools, the motives and the psychopaths to do it.

“For more than forty years we have known that like DNA, RNA is chemically modified, with evidence of RNA modifications identified from viruses to Arabidopsis, mouse and man. Characterisation of highly abundant modified tRNA and rRNA first informed us of the plethora of structural and functional roles for modified RNA. “
HeatherCoker, GuifengWei, NeilBrockdorff – Department of Biochemistry, University of Oxford

also see:
WE WRITE NEW DNA USING RNA ONLY – STAR SCIENTIST FINANCED BY EPSTEIN, DARPA AND SCHWAB’S WYSS INST.
MICROSOFT, 2016: “WE CAN PROGRAM COMPLEX BEHAVIORS USING DNA”. 3-STRAND DNA CONFIRMED
To be continued?
Our work and existence, as media and people, is funded solely by our most generous supporters. But we’re not really covering our costs so far, and we’re in dire needs to upgrade our equipment, especially for video production.
Help SILVIEW.media survive and grow, please donate here, anything helps. Thank you!
! Articles can always be subject of later editing as a way of perfecting them